Processing DICOM file in Python with Pydicom
DICOM files are very common in medical imaging projects. They provide a data type that integrates the image pixel data and image information data. Pydicom is quite a convenient package to read and process DICOM files in Python.
In this post, I will show a few simple examples, and explain a bit about the compressed pixel data issue with Pydicom.
Read and display a DICOM file
To install Pydicom, simply use:
pip install pydicom
Now let’s try to read some DICOM files and dispplay it.
First we import some nessassry packages.
import pydicom
import numpy as np
import matplotlib.pyplot as plt
import os
Curenttly I have a local folder named Patient1 contain some chest CT data.
path = 'Patient1/'
#loading the DICOM file names
files_list = sorted(os.listdir(path))
print(files_list)
['000001.dcm', '000002.dcm', '000003.dcm', '000004.dcm', '000005.dcm', '000006.dcm', '000007.dcm', '000008.dcm', '000009.dcm', 'I0001283.dcm']
Now we can see there are 10 DICOM images of chest CT data in this folder.
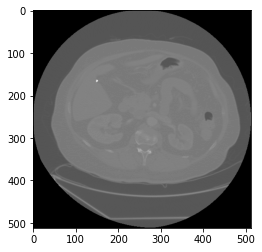
We can now try to read one of the image. And check what’s inside its pixel array.
sampledicom = pydicom.dcmread(path + files_list[0])
sampledicom.pixel_array
array([[-2000, -2000, -2000, ..., -2000, -2000, -2000],
[-2000, -2000, -2000, ..., -2000, -2000, -2000],
[-2000, -2000, -2000, ..., -2000, -2000, -2000],
...,
[-2000, -2000, -2000, ..., -2000, -2000, -2000],
[-2000, -2000, -2000, ..., -2000, -2000, -2000],
[-2000, -2000, -2000, ..., -2000, -2000, -2000]], dtype=int16)
There are a few ways to visualize the pixel_array data. One way is using the matplotlib package. Check out other methods here.
plt.imshow(sampledicom.pixel_array, cmap=plt.cm.gray)
<matplotlib.image.AxesImage at 0x21ea9e50df0>
View some other elements using Pydicom
One benefit of DICOM files is to have pixel and medical-related information in one single file. Thus, we can access some crucial information for medical image processing from any DICOM files.
Let’s start by looking at the meta data of a DICOM file.
sampledicom.file_meta
(0002, 0000) File Meta Information Group Length UL: 206
(0002, 0001) File Meta Information Version OB: b'\x00\x01'
(0002, 0002) Media Storage SOP Class UID UI: CT Image Storage
(0002, 0003) Media Storage SOP Instance UID UI: 1.3.6.1.4.1.14519.5.2.1.6279.6001.278631323634593956582096991980
(0002, 0010) Transfer Syntax UID UI: Explicit VR Little Endian
(0002, 0012) Implementation Class UID UI: 1.3.6.1.4.1.22213.1.143
(0002, 0013) Implementation Version Name SH: '0.5'
(0002, 0016) Source Application Entity Title AE: 'POSDA'
You can also get other information for the specific DICOM file. For example, you can get the CT image slice thickness from the DICOM file as well.
print(f"Slice Thickness: {sampledicom.SliceThickness}")
Slice Thickness: 1.250000
Reading compressed pixel data in DICOM files
Pixel data are normally stored in DICOM files after compression. And to be able to read more types of data, some other packages might be needed to be able to read your own DICOM files data.
You probably noticed that in the first section, when I printed the file names. There is one file I0001283.dcm that is different from other files. This file is actually from another CT series, which I can compare the file differences between two CT series.
We can check the UID and BitsAllocated information in a few lines of codes.
print('The DICOM file we seen:')
print(sampledicom.file_meta.TransferSyntaxUID)
print(sampledicom.BitsAllocated)
specialdicom = pydicom.dcmread(path + files_list[9])
print('The other DICOM file from another series:')
print(specialdicom.file_meta.TransferSyntaxUID)
print(specialdicom.BitsAllocated)
The DICOM file we seen:
1.2.840.10008.1.2.1
16
The other DICOM file from another series:
1.2.840.10008.1.2.4.51
16
You can check more details about compressed pixel data and the supported UID information on this page.
We can find out that the first CT data we used is supported by pydicom without any third-party packages, which is great. But for the second file, we can see pydicom will need additional packages to be able to read the pixel information in the DICOM file.